The MALDI Biotyper identifies microorganisms by analyzing their intrinsic proteins using mass spectrometry.
An individual colony from an overnight culture grown on an agar plate is picked using a wooden toothpick and transferred to a MALDI target. This sample is allowed to dry, and a microliter of a matrix solution is added to the sample. The organic solvent in the matrix solution extracts proteins from the microorganisms. The extracted proteins are mainly ribosomal proteins, present in high concentrations. Once the matrix has crystallized, sample preparation is complete and the samples are ready for analysis. For some microorganisms, it is necessary to perform a more extensive extraction, which adds another 10-15 minutes to the sample preparation procedure.

Samples are ready to be analyzed. (MSP 96 target polished steel; Bruker Part No: 8280800)
In our facility, the analyses are carried out on the Bruker Ultraflextreme MALDI TOF/TOF mass spectrometer. Each MALDI mass spectrum is a species-specific molecular fingerprint, a mass and intensity distribution of peaks corresponding to mainly ribosomal proteins.
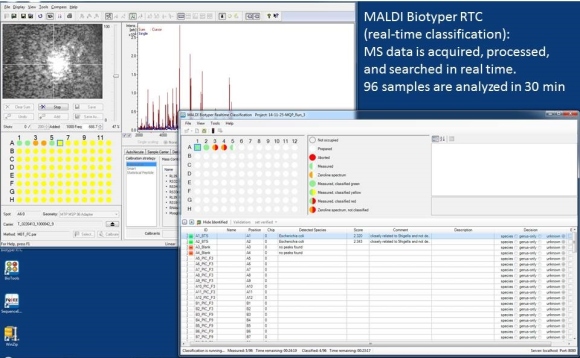
Microorganisms are identified in real time as soon as data are collected from each individual target position.
Identification of an unknown microorganism is based on comparing the experimental molecular fingerprint with 6903 fingerprints in the Biotyper library and assigning a score which represents the probability that the match is correct.

Off-line classification software provides additional options for working with data such as creating new library entries, re-organizing libraries, viewing mass spectra, or assessing quality of match.
The algorithm used to compare patterns computes three separate values for three fundamental characteristics of the sample spectra and the reference spectra: the number of signals in the reference spectrum that have a closely matching partner in the unknown spectrum; the number of signals in the unknown spectrum that have a closely matching partner in the reference spectrum; and the intensity symmetry of the matching pairs. The maximum obtainable score is 3, and the score values greater or equal to 2 are considered as a probable identification.

Each mass spectrum is converted into a histogram which represents the mass and intensity ‘fingerprint’.
Biotyper software allows building spectral libraries of any type of samples as long as the samples are amenable to MALDI-TOF MS and produce unique MS fingerprints. If you are interested in learning more about MALDI MS and the Biotyper, please contact Tatiana.
The acquisition of MALDI Biotyper software was possible through collaboration with Dr. Bhushan M. Jayarao, Director of Penn State Animal Diagnostic Laboratory, Professor of Veterinary and Biomedical Sciences.
Featured image credit: Rocky Mountain Laboratories, NIAID, NIH
Scanning electron micrograph of Escherichia coli, grown in culture and adhered to a coverslip






