If you have never done in-gel protein digestion, this tutorial is for you! The protocol is really simple and does not require any specialized equipment. Even if your gel has been sitting in the fridge for a month or two, it should still work, but no mold, please! First order of business is to choose a band and cut it out. You will need a clean blade, a clean surface (a clean glass plate or a clean transparency sheet), a clean microcentrifuge tube, and a pair of clean gloves. Everything should be clean: the goal is to minimize contamination of interesting proteins by uninteresting keratins.
1. Place the gel on a glass plate, blot excess water with a clean paper tissue. Select a band and cut out only the stained portion of the gel. Avoid unstained area: it will sponge up a protease solution, giving nothing interesting in return.
2. Dice the band into 1-mm (1/16”) sections; this will help the protease reach more protein inside the gel.
3. Place the gel cubes in a clean microcentrifuge tube, cover with water to prevent them from drying out, and label the tube with no more than 5 characters. For example, use your initials and numbers.
4. Include a positive control! There is no charge for analyzing a control! Cut out a known protein band following steps 1, 2, and 3.

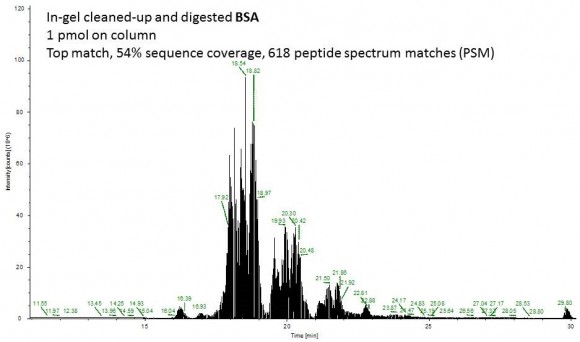
The in-gel digestion protocol can be found on Thermo website along with the product numbers for all necessary reagents.
Washing the gel
You will need 50% acetonitrile (ACN) solution containing 8 mg/mL ammonium bicarbonate (wash solution). Add 0.05 – 0.1 mL of this solution per gel band, enough to completely cover the gel. Incubate at 37 C for 15-20 min, discard the solution. Repeat two more times. At this point, all or most of the blue color should be gone. If you are destaining SYPRO, there is no easy way to tell whether it is all gone, of course.
Reducing disulfides and alkylating Cys residues
You will need
- 5 mM TCEP (Tris(2-carboxyethyl)phosphine hydrochloride) solution in the 8 mg/mL ammonium bicarbonate. I make 100 mM (29 mg/mL) stock solution in water and store it in the fridge up to a month. Dilute this stock 20x (e.g. 50 uL 100 mM TCEP + 950 uL ammonium bicarbonate solution).
- Fresh 100 mM IAA (iodoacetamide) solution in the 8 mg/mL ammonium bicarbonate. This is approximately 18 mg/mL IAA solution.
Cover the gel slices with 5 mM TCEP and incubate 10 min at 60 C. Remove the TCEP solution and cover the gel with 100 mM IAA solution. Incubate at 37 C for 15 min with occasional shaking, preferably protected from light. Room temperature will also work, but give it extra 10-15 min. Discard the IAA solution and wash the gel with the wash solution three times to remove IAA and TCEP. (In the Pierce protocol, you might notice that 100 mM IAA requires 9.3 mg in 1 mL, while MW of IAA is 184.9. It is a typo. Either way: 50 mM, 9.3 mg/mL or 100 mM, 18 mg/mL, will work as there will be a large molar excess of IAA compared to Cys. Shrink the gel by covering it with ACN and incubating at r.t. for 15 min or until the gel turns white and brittle. Remove ACN and allow the gel to air dry for 15 min at 37 C (or longer at room temperature).
Digestion
Make 1 mg/mL trypsin stock solution in 50 mM acetic acid or 1 mM hydrochloric acid. This solution can be aliquoted and stored in a freezer for months. Dilute trypsin to 0.01 mg/ml (1:100 dilution) with 8 mg/mL ammonium bicarbonate. Add 50 uL of this solution per gel band and incubate at 37 C 8-24 hours. If you are working with a large piece of gel or several bands combined in one tube, make sure that after the gel re-hydrates in the enzyme solution, it is completely covered. Add more enzyme solution if necessary.
Extracting the peptides
You will need 50% ACN solution containing 0.1% formic acid (FA). If you don’t have formic acid in your lab, let me know – we will share ours with you. Transfer the digest solution to a new tube. Extract each gel band with 50 uL of 50% ACN/0.1% FA (more, if working with a large volume of gel) by incubating at 37 C for 15 min. Transfer this solution to the new tube with the digest solution. Repeat 2 more times. Evaporate the combined extracts in a vacuum concentrator (e.g. Speedvac). Please note, if you are extracting large peptides (>5000 Da), using a sonic bath may be a better option than incubation. Also, in addition to the three 50% ACN extractions, you can use 100% acetonitrile for the final extraction. Submit your dried samples along with a picture of the gel so that I can estimate how much of each sample to use for analysis, and don’t forget to fill out the Protein ID Request form
As always, let me know if you have any questions!