Elvira-Matelot E, Hachet M, Shamandi N, Comella P, Saez-Vasquez J, Zytnicki M, Vaucheret H
The Arabidopsis RTL2 is an RNASE THREE LIKE protein with one RNAseIII domain and two dsRNA-binding domains. Its transient over-expression in plants is known to enhance the production of exogenous siRNAs. Here the authors investigate its role in the production of endogenous siRNAs.
The ectopic expression of RTL2 stimulates the production of siRNAs from artificial and natural Inverted Repeats constructs. Both its domains are necessary for the RTL2-dependent production of siRNAs from dsRNAs. Contrarily, in other cases the over-expression of RTL2 reduces the production of siRNAs from dsRNAs. The opposite effect of RTL2 on dsRNA substrates likely depends on the structure and/or sequence of the dsRNAs.
Interestingly, the over-expression of RTL2 also stimulates the production of RNA molecules larger than 24 nts from artificial and natural Inverted Repeats constructs. The authors also demonstrate that RTL2 cannot substitute for the function of DCL2, DCL3 and DCL4. The suggested hypothesis is that RTL2 could process dsRNAs into RNA molecules longer than 24 nts, which could subsequently be processed by DCL proteins into siRNAs. In some cases the cleavage of RTL2 results in a better processing by DCLs, in some other cases it results in a worse processing by DCLs.
By a sliding window approach combined with the analysis of reads at each position of the genome, a total of 481 sRNA loci are found to be differentially expressed in the rtl2 mutant compared to wt: 183 are RTL2-dependent loci, 298 are RTL2-sensitive loci. These RTL2-targeted sRNA loci produce siRNAs that are mostly dependent on DCL2DCL3DCL4, Pol IV, Pol V and DRM2, indicating the involvement of these sRNA loci in the RdDM process.
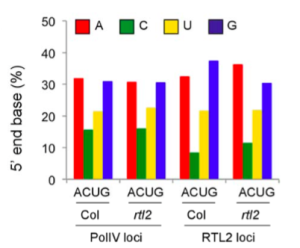
Recent works reported that siRNA precursors, named P4R2 RNAs (Pol IV- and RDR2-dependent), preferentially start with an A or a G, and preferentially end with a U (Blevins et al. 2015 and Zhai et al., 2015). The authors found that the RTL2 loci show different 5’ and 3’ nucleobase preferences compared to the total Pol IV loci previously identified. This tendency is reverted to the same behavior observed for the total Pol IV loci in the rtl2 mutant, suggesting that RLT2 modifies the 5’ and 3’ end compositions of the target loci, making them similar to those of the total P4R2 RNAs.
RTL2 targets mainly TEs and intergenic regions but also protein-coding genes, influencing the DNA methylation and mRNA expression level of the target loci.
The biogenesis of siRNAs is not yet completely understood and these findings suggest that the siRNA precursors might undergo multiple successive processings operated by different proteins. The one precursor one siRNA model might not be valid for all sRNA genes, leaving the question: what are the genetic/epigenetic features of the sRNA genes that differentiate their sRNA precursors production and processing?
Alice