Electrospray ionization (ESI) and matrix-assisted laser desorption ionization (MALDI) are both competitive ionization processes in which a more abundant and/or more readily ionizable species will always win. Unless you are studying ionization of Tris, PEG, EDTA or CHAPS in the presence of small amounts of protein, the protein solution submitted for MS analysis should be essentially free of salts, detergents, chaotropic reagents, glycerol and other uninteresting interferences.
“So why not take a 10 mg/mL protein solution in a ‘mild buffer’ and dilute it to the required 0.1 mg/mL; the buffer concentration will be 100 times lower, and it is as good as desalted, right?” Wrong and here’s why:
Dilution is not a solution
Let’s say you have a 10 mg/mL solution of a 10 kDa protein in 10 mM Tris buffer, ‘mild’ enough. This means, you have a 10-fold molar excess of Tris compared to the protein. No matter how much you dilute this solution, the molar ratio of Tris to protein will remain 10:1 while it should be 1:50 or less.
For practical purposes, ‘essentially salt-free’ means that the total molar concentration of interfering species is much lower than that of the protein, e.g. at least 50-fold lower.
Clean-up strategies
There are many ways to clean up a protein solution. In each case, the choice of a protocol will depend on the properties of interfering species and on the size and properties of the protein. For example, detergents can be removed using specialized detergent removal spin columns. PEGs and other charge-neutral contaminants can be washed away by loading the protein onto an ion exchange medium, eluting the protein with a high-salt buffer, and desalting the eluate. Buffer salts, urea, and other small molecules can be removed by either a desalting spin column packed with size-exclusion resin or a centrifugal filter with an appropriate molecular-weight-cut-off (MWCO) membrane. If you need an advice on your protein sample clean-up, I am always happy to help; you know where to find me!
Desalting using centrifugal filters
Centrifugal filters with different MWCO membranes are probably the easiest way to desalt a small volume of protein solution, although there are a few caveats. For example, the regenerated cellulose Ultracel membranes might retain cellulose-binding proteins; in this case, the low protein binding Omega membranes from PALL would be a better choice.
How many spin-dilute cycles are enough to ‘completely’ desalt a protein solution? This is one of those annoying situations where the old trusty triplicate doesn’t always work. Here’s my handy formula to help me decide when to stop desalting and start analyzing:
Example 1.
Let’s use our 10 mg/mL 10 kDa protein in 10 mM Tris buffer and a 3 kDa MWCO Amicon centrifugal filter as an example. The filter has a 0.5 mL capacity, and we can load 50 uL of the protein solution and dilute it with 450 uL of water. After a 30-min centrifugation, the volume is 50 uL, but the concentration of Tris is now 10% (or 0.1) of the initial while the protein concentration is essentially unchanged. So our D, dilution factor for Tris after each dilution+spin is 0.1; our goal, M final is 0.01 mM Tris (100:1 protein:Tris); and our M initial is 10 mM Tris. To find the number of dilute+spin cycles, dust off your scientific calculators:
 After 3 dilute/spin repeats, we will achieve our acceptable concentration of Tris, 100 times lower than the protein concentration.
After 3 dilute/spin repeats, we will achieve our acceptable concentration of Tris, 100 times lower than the protein concentration.
Example 2.
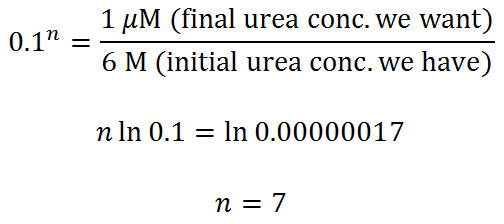
Let’s throw in some urea, dilute the protein to 0.1 mM, and do our calculation again. How many steps (n = ?) do we need to get from 6 M to 1 uM urea (100:1 protein-to-urea ratio) using the same spin column as before?
You probably can get away with 6 dilute/spin repeats, but 7 would be better.
For those of you who hate equations, here’s a picture that illuminates the desalting process using the first example.
“What if my protein is unstable without a buffer?”
In that case, it may not be amenable to the direct ESI MS. However, before throwing up your hands, try to desalt your protein. It might be stable in water at micromolar concentration long enough to be analyzed – I am speaking from experience. You also have an option of using LC ESI MS or submitting your partially desalted protein for MALDI TOF MS.