Tag Archives: in-gel digestion
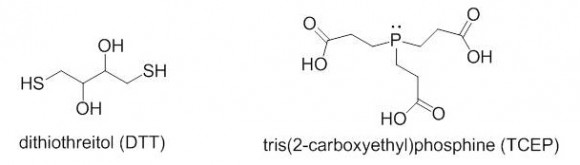
TCEP or DTT?
Which reagent is better for reducing disulfides, DTT or TCEP? The two reagents are quite different in their reactivity, stability towards oxidation, reaction mechanisms, and other categories.
DTT is a thiol-containing reagent, and this must be considered in applications involving thiol labeling. TCEP is charged in solution and should not be used in isoelectric focusing. Aqueous solutions of TCEP are quite acidic (pH 2-3).
TCEP HCl is odorless, air-stable crystalline solid, soluble in water at a > 1 M concentrations. It reduces disulfides at room temperature in < 5 min in dilute solutions (5 -50 mM). There is no need to remove TCEP prior to the use of sulfhydryl-reactive labels or crosslinkers. TCEP is selective toward disulfides, and is reactive at a broad pH range.
For those who love Chemistry:

Reduction of disulfide with TCEP. First step is rate-determining, kinetic rather than thermodynamic control.
You can learn more about DTT and TCEP from this article: “A Comparison between the Sulfhydryl Reductants Tris(2-carboxyethyl)phosphine and Dithiothreitol for Use in Protein Biochemistry”, Elise Burmeister Getz et al. Analytical Biochemistry 273, p. 73–80 (1999)
Ammonium bicarbonate or triethylammonium bicarbonate?
This was a question from one of my blog’s secret readers. Actually, most of the time I feel like I am talking to myself: “Hey Tania, how do you prepare a protein sample for proteolysis?” “Well, Tania, let me show you in a step-by-step tutorial.” No comments, no questions, no pointing out typos, no “thank you, Tania, but there’s a better way to do this”?
Oh well, back to ammonium bicarbonate. This is a volatile salt which breaks down to ammonia, carbon dioxide, and water. Volatile salts are the only salts compatible with MS. Aqueous solutions of ammonium bicarbonate (0.01 – 0.1 M) have pH around 8, the optimal pH for trypsin activity. Ammonium bicarbonate competes with basic amino acids for Coomassie dye, which makes it a great de-staining reagent for the in-gel digestion procedure. All this goodness comes at a very reasonable price – what not to like? Another ammonium salt, triethylammonium bicarbonate (TEAB), is more volatile than ammonium bicarbonate; it is also more expensive. TEAB is a buffer of choice for LC-MS applications: TMT (iTRAQ) amine-reactive labeling, ion-exchange chromatography, protein solubilization (when neutral and acidic pH is undesirable), in-gel digestion, etc.
Detergent removal 1: Gel-assisted proteolysis
As promised, here’s a straightforward way to remove detergents, urea, and other LC-MS incompatible nasties from small-volume samples. The literature calls it ‘gel-assisted’ proteolysis. The idea is to entrap the protein solution in a polyacrylamide gel matrix, wash out detergents, salts, and chaotropic agents, and perform in-gel digestion. This technique works great for membrane proteins which are notoriously difficult to dissolve, and it is quite useful for any protein sample clean-up.
For my little demo, I used a 1 mg/mL BSA solution in 2% SDS. The disulfides were reduced with TCEP and alkylated with IAA, after which the protein solution was very thoroughly mixed with a 30% T acrylamide monomer solution. I quickly added 10% APS and TEMED and immediately vortexed and centrifuged this mixture so that the liquid is collected at the bottom of the tube. The polymerization time is very short, a minute or two! I left it to completely polymerize for another 20 min.
Using scalpel I removed the gel plug from the tube and diced it into small pieces. After 6 washes with 8 mg/mL ammonium bicarbonate in 50% acetonitrile, I dried the gel pieces in neat acetonitrile, removed the acetonitrile and added trypsin (see the in-gel digestion tutorial for details).
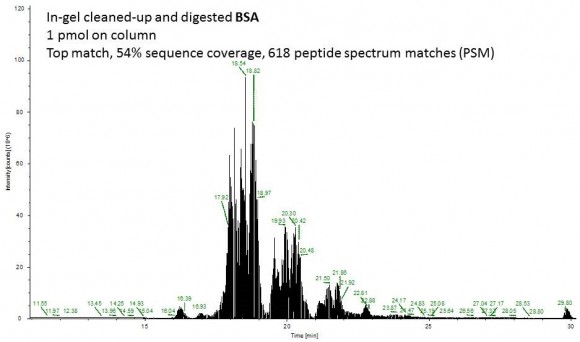
Fast forward to the MS analysis: Since the original BSA solution was very concentrated, I dissolved the peptides in 540 uL of mobile phase and injected 1 uL of this solution (55 ng total protein on column or approximately 1 pmol).
Second example shows an ion chromatogram from a 10-uL IP eluent containing 2% detergent which I cleaned up and digested using this technique.
As always, let me know if you have questions!
References:
doi: 10.1074/mcp.M500138-MCP200
doi: 10.1074/mcp.M800068-MCP200
Extracting large peptides from gel after digestion
First, let’s define ‘large’. The Proteome Discoverer sets the upper limit for a precursor ion at 10,000 Da. This means anything bigger than 10 kDa will not be considered even if it’s present in the MS data, and a different software package ($$) will be required to analyze the high MW data. Clearly, proteolytic peptides with MW > 10 kDa will not be very useful for protein identification. I suggest using a different enzyme or a combination of enzymes. I have seen tryptic peptides up to 7 kDa in some in-gel digested samples, so apparently some large peptides do come out of gel.
Next, a large peptide’s physico-chemical properties (e.g. hydrophobicity, pI, hydrodynamic radius) must be considered as they will affect the extraction efficiency. If the peptide’s properties are known, the extraction solvent composition and pH can be adjusted to improve the peptide’s solubility.
Finally, let’s consider the gel from which the large peptides need to come out. Obviously, it will be easier to get the large peptides out of a 4 %T gel than out of a 20 %T one. Soaking a gel piece in deionized water and then freezing it should crash enough pores in the gel to improve the extraction of large peptides (water expands as it freezes). Additionally, the gel could be ‘squeezed out’ a few times by changing extraction solvent from neat acetonitrile to an aqueous mixture. The gel piece will shrink in acetonitrile expelling the peptide solution. Re-hydrating the gel and then shrinking it again in acetonitrile will ‘squeeze out’ more of the digest.
Using elevated temperature (50 C), vortex mixer, and/or ultrasonic bath should all improve the extraction. Use common sense: 50C and a high pH buffer is not a good idea for the phosphopeptide extraction. Another word of caution: don’t get carried away. Three extraction steps should be enough. If you end up with a large volume (e.g. more than 0.5 mL), the benefits of a thorough extraction might become negated by the losses due to dilution. Peptides and proteins tend to adhere to the polypropylene tubes. A large volume of a dilute peptide solution presents a large surface area for the peptides to adsorb.
What to do if this doesn’t work? You can try in-solution digestion. If the mixture is too complex and a PAGE step is necessary, you can try electroeluting the protein(s). Intact proteins electroeluted from gel bands can be buffer-exchanged using small-volume 3,000 Da MWCO spin columns and proteolyzed in solution.
Tutorial: In-gel digestion
If you have never done in-gel protein digestion, this tutorial is for you! The protocol is really simple and does not require any specialized equipment. Even if your gel has been sitting in the fridge for a month or two, it should still work, but no mold, please! First order of business is to choose a band and cut it out. You will need a clean blade, a clean surface (a clean glass plate or a clean transparency sheet), a clean microcentrifuge tube, and a pair of clean gloves. Everything should be clean: the goal is to minimize contamination of interesting proteins by uninteresting keratins.
1. Place the gel on a glass plate, blot excess water with a clean paper tissue. Select a band and cut out only the stained portion of the gel. Avoid unstained area: it will sponge up a protease solution, giving nothing interesting in return.
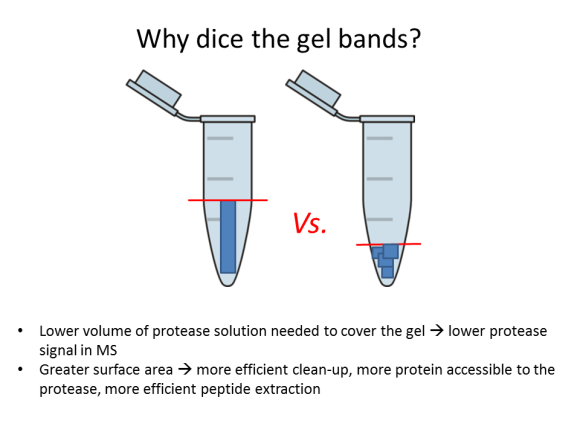
2. Dice the band into 1-mm (1/16”) sections; this will help the protease reach more protein inside the gel.
3. Place the gel cubes in a clean microcentrifuge tube, cover with water to prevent them from drying out, and label the tube with no more than 5 characters. For example, use your initials and numbers.
4. Include a positive control! There is no charge for analyzing a control! Cut out a known protein band following steps 1, 2, and 3.

The in-gel digestion protocol can be found on Thermo website along with the product numbers for all necessary reagents.
Washing the gel
You will need 50% acetonitrile (ACN) solution containing 8 mg/mL ammonium bicarbonate (wash solution). Add 0.05 – 0.1 mL of this solution per gel band, enough to completely cover the gel. Incubate at 37 C for 15-20 min, discard the solution. Repeat two more times. At this point, all or most of the blue color should be gone. If you are destaining SYPRO, there is no easy way to tell whether it is all gone, of course.
Reducing disulfides and alkylating Cys residues
You will need
- 5 mM TCEP (Tris(2-carboxyethyl)phosphine hydrochloride) solution in the 8 mg/mL ammonium bicarbonate. I make 100 mM (29 mg/mL) stock solution in water and store it in the fridge up to a month. Dilute this stock 20x (e.g. 50 uL 100 mM TCEP + 950 uL ammonium bicarbonate solution).
- Fresh 100 mM IAA (iodoacetamide) solution in the 8 mg/mL ammonium bicarbonate. This is approximately 18 mg/mL IAA solution.
Cover the gel slices with 5 mM TCEP and incubate 10 min at 60 C. Remove the TCEP solution and cover the gel with 100 mM IAA solution. Incubate at 37 C for 15 min with occasional shaking, preferably protected from light. Room temperature will also work, but give it extra 10-15 min. Discard the IAA solution and wash the gel with the wash solution three times to remove IAA and TCEP. (In the Pierce protocol, you might notice that 100 mM IAA requires 9.3 mg in 1 mL, while MW of IAA is 184.9. It is a typo. Either way: 50 mM, 9.3 mg/mL or 100 mM, 18 mg/mL, will work as there will be a large molar excess of IAA compared to Cys. Shrink the gel by covering it with ACN and incubating at r.t. for 15 min or until the gel turns white and brittle. Remove ACN and allow the gel to air dry for 15 min at 37 C (or longer at room temperature).
Digestion
Make 1 mg/mL trypsin stock solution in 50 mM acetic acid or 1 mM hydrochloric acid. This solution can be aliquoted and stored in a freezer for months. Dilute trypsin to 0.01 mg/ml (1:100 dilution) with 8 mg/mL ammonium bicarbonate. Add 50 uL of this solution per gel band and incubate at 37 C 8-24 hours. If you are working with a large piece of gel or several bands combined in one tube, make sure that after the gel re-hydrates in the enzyme solution, it is completely covered. Add more enzyme solution if necessary.
Extracting the peptides
You will need 50% ACN solution containing 0.1% formic acid (FA). If you don’t have formic acid in your lab, let me know – we will share ours with you. Transfer the digest solution to a new tube. Extract each gel band with 50 uL of 50% ACN/0.1% FA (more, if working with a large volume of gel) by incubating at 37 C for 15 min. Transfer this solution to the new tube with the digest solution. Repeat 2 more times. Evaporate the combined extracts in a vacuum concentrator (e.g. Speedvac). Please note, if you are extracting large peptides (>5000 Da), using a sonic bath may be a better option than incubation. Also, in addition to the three 50% ACN extractions, you can use 100% acetonitrile for the final extraction. Submit your dried samples along with a picture of the gel so that I can estimate how much of each sample to use for analysis, and don’t forget to fill out the Protein ID Request form
As always, let me know if you have any questions!