Cassie McMillan
2016-12-05
This vignette is designed to give an introduction of how to perform network analysis in R using the statnet package. Statnet is a suite of several network-related R packages including sna, network, ergm, and tergm. The statnet website includes a complete list of the included packages. While one of the main focuses of statnet is the statistical modeling of networks, this vignette will mainly focus on data handling, calculating descriptives, and visualization. At the end, I’ll provide resources you can check out if you’re interested in learning more about more advanced functions in statnet. Also, I’ll quickly note that there exist other network analysis R packages, most notably igraph. All these packages are able to do the more basic stuff there are some differences that are worth checking out.
Getting Started
Like any R package, it’s first necessary to install and access it. The package can easily be installed through CRAN.
install.packages("statnet")
library(statnet)For this vignette, I’ll be using data that David Krackhardt collected from managers in a company. There are two csv files containing the data. The first krack_advice is an adjacency matrix indicating who gives advice to who. The second krack_attributes is a list of attributes for each of the managers such as age and tenure. After setting your working directory, load in the data like this:
advice <- read.csv("Krack_Advice.csv", header=T, row.names=1, check.names=FALSE)
#include header = T and row.names = 1 to account for the node labels
att = read.csv("Krack_Attributes.csv", header=T)
advice
head(att)Data Management
First, I’ll go over how to manage network data using statnet. To use most of statnet’s functions it is necessary to first convert the adjacenecy matrices into network objects. This can be done in the following code:
advice.m = as.matrix(advice) # tell R data should be understood as a matrix
advice.n =network(advice.m, matrix.type="adjacency",directed=TRUE) # transforms into a network object
advice.mThe argument matrix.type specifies that we are putting in an adjacency matrix, you can also read in edge lists. Also in the network command, you can specify whether the matrix is directed or undirected, if it’s a bipartite network, if self-loops are allowed, etc.
Next, you need to link the attribute data with the network object. To do this it’s necessary that the nodes in the network object and attribute data be in the same order. Then, there are two ways to link the data. First, you can go in and add the attributes in one at a time. Alternatively, you can add the attributes in when you make the network option. It’s also possible to add in edge attributes, but the current data set does not have any of these.
#check if nodes are in the same order
att$ID==rownames(as.sociomatrix(advice.n))
#option 1
advice.n %v% "dept" <- att$DEPT
advice.n
#option 2
advice.n = network(advice.m, matrix.type = "adjacency", directed = T, vertex.attr = att)
advice.nIf you need to, you can easily add in new edges to the network object or delete existing edges. You can also check whether edges exist between pairs.
advice.n[2,1] <- 1 #adds an advice tie from node 2 to node 1
advice.n[2,1] <- 0 #removes an advice tie from node 2 to node 1
advice.n[1,2] #does node 1 send an advice tie to node 2?Reading in your network data is pretty simple if your network data are already in adjacency matrices. However, network data can come in a variety of formats, including adjacency lists and edge lists. If your data are in this form, then there’s a lot of code out there that you can use to transform your data into adjacenecy matrices, which tend to be much easier to work with. In the past, I’ve used code from this blog to transform other data formats into adjacenecy matrices.
Descriptives
Now I’m going to present a couple descriptive measures that are available from the statnet package. First, here’s a bunch of functions that calculate network-level descriptives:
summary(advice.n) #summary of network object, also provides an edge list
network.size(advice.n) #number of nodes in the network
network.dyadcount(advice.n) #number dyads that exist in network (n*(n-1))
network.edgecount(advice.n) #number of edges present in the network
gden(advice.n) #network density (edge count/dyad count)
grecip(advice.n, measure = "dyadic") #proportion of symmetric dyads
gtrans(advice.n, measure = "weak") #proportion of transitive triads
symmetrize(advice.n, rule = "weak") #symmetrize so i<->j iff i->j OR i<-j
symmetrize(advice.n, rule = "strong") #symmetrize so i<->j iff i->j OR i<-j
dyad.census(advice.n) #MAN dyad census
triad.census(advice.n) #standard directed triad census
kpath.census(advice.n, maxlen=3, tabulate.by.vertex=FALSE) # Count paths of length <=3
kcycle.census(advice.n, maxlen=3, tabulate.by.vertex=FALSE) # Count cycles of length <=3
clique.census(advice.n, tabulate.by.vertex=FALSE, enumerate=FALSE) # counts of cliques by sizeAnd here are some node-level descriptive statistics:
degree(advice.n, cmode="indegree") #indegree, number of nominations received
degree(advice.n, cmode="outdegree") #outdegree, number of nominations sent
degree(advice.n) #total degree (sent+received)
betweenness(advice.n) #betweenness
closeness(advice.n) #closeness
isolates(advice.n) #lists the isolates in the graph
geodist(advice.n) #gives number and lengths of all geodesics (shortest paths) between all nodes Visualization
The statnet package also allows for you to easily visualize your network graphs with the gplot function.
gplot(advice.n)There are a lot of interesting arguments that you can add into the gplot function. You can add in vertex labels and change the size and color of these labels:
gplot(advice.n, displaylabels=TRUE,
label.cex=.75, label.col="black")For directed graphs, you can turn off the arrows. This tends to be especially helpful for large graphs with a lot of nodes and edges.
gplot(advice.n, displaylabels=TRUE,
label.cex=.75, label.col="black", usearrows = FALSE)It’s easy to differentiate nodes based on their attributes. You can color the nodes based on their attributes. The code below colors attributes based on their department. Each color represents a different department. I also include code here for adding a legend that tells us what each color represents.
gplot(advice.n, displaylabels=TRUE,
label.cex=.75,label.col="black",vertex.col=att$DEPT)
legend("bottomleft",fill=0:4,legend=paste("DEPT",0:4),cex=0.75)
Here, I changed the shape of the nodes based on the level of their positions. You change the shape of the nodes by specifying the number of sides you want the shape to have using the vertex.sides argument. For instance, vertex.sides = 4 will result in squares. If you want circles, set vertex.sides = 50.
gplot(advice.n, displaylabels=TRUE,
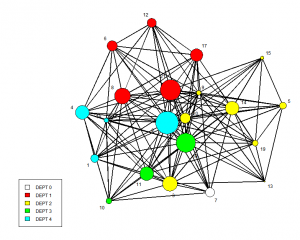
label.cex=.75,label.col="black",vertex.cex = 2, vertex.sides=(att$LEVEL+2))You can also change the size of the nodes based on an attribute. In the examples below, I do this for both tenure of the employees and the indegree (nominations received) by each employee. I divide the size values by 6 so they can be reasonably scaled. In the graph shown below, the nodes have been sized according to indegree. Larger nodes received more advice nominations.
#sized by tenure
gplot(advice.n, displaylabels=TRUE,
label.cex=.75,label.col="black", vertex.cex = (att$TENURE/6))
#sized by indegree
gplot(advice.n, displaylabels=TRUE,
label.cex=.75,label.col="black", vertex.cex = (degree(advice.n, cmode="indegree")/6))As a default, gplot uses the Fruchterman Reingold algorithm to lay out the nodes. However, you can change this as well. For instance, you can lay out the nodes using MDS or in a circle.
gplot(advice.n, displaylabels=TRUE,
label.cex=.75,label.col="black", mode = "mds") #multi-dimension scaling
gplot(advice.n, displaylabels=TRUE,
label.cex=.75,label.col="black", mode = "circle") #circleIf you like a layout, you can save the coordinates of it and then reapply these coordinates later to preserve your same layout.
coordinates <- gplot(advice.n, displaylabels=TRUE,
label.cex=.75,label.col="black")
coordinates #prints the coordinates
#applying saved coordinates to a new graph
gplot(advice.n, displaylabels=TRUE,
label.cex=.75,label.col="black", coord = coordinates)Note that you can also use the coord argument to make up your own coordinates. The Krackhardt data we’ve been working with doesn’t have any isolates (e.g. those who neither send nor receive ties), but when we do have data with isolates these can often get annoying when visualizing. As a result, you can plot a graph without displaying isolates by including the following argument: displayisolates=FALSE.
Furthermore, gplot does include an interactive function where you can move around the positioning of vertices yourself until you find a display that you like.
gplot(advice.n, displaylabels=TRUE,
label.cex=.75,label.col="black", interactive=TRUE)There are also several other arguments that you can include to make graphs that are both pretty and interesting.
palette(rainbow(6))
gplot(advice.n, displaylabels=TRUE,
label.cex=.75,label.col="black",
usecurve==TRUE, vertex.col=att$DEPT,
vertex.cex = (degree(advice.n, cmode="indegree")/7),
edge.col = "black", usearrows = FALSE,
edge.curve = 0.5, vertex.border = "black")
legend("bottomleft",fill=0:4,legend=paste("DEPT",0:4),cex=0.75)Statistical Modeling
As mentioned previously, the statnet package also includes a lot of functions for statistically modeling networks. This includes QAP correlations, MRQAP, ERGMs, and TERGMs/STERGMs. An in-depth discussion of these packages is beyond the scope of this vignette, but here are a couple additional resources that go into more detail about these functions. I’ve found these to be helpful in the past at explaining how these functions work: