Overview
 Folding of RNA in the cell is not well understood nor has it been integrated into a cohesive mechanistic framework. We are taking two approaches to understanding RNA folding in vivo: (1) Simulating in vivo conditions and examining the RNA folding pathways and the evolutionary driving forces for these, and (2) Determining RNA folding transcriptome-wide in living organisms (plants, bacteria, and archaea) and evaluating implications of this folding on gene regulation and RNA processing. The former project has the advantage that one can precisely control experimental variable such as crowding, cosolutes, and ionic conditions on RNA folding, while the latter allows observations to be made made directly in a living organism.
Folding of RNA in the cell is not well understood nor has it been integrated into a cohesive mechanistic framework. We are taking two approaches to understanding RNA folding in vivo: (1) Simulating in vivo conditions and examining the RNA folding pathways and the evolutionary driving forces for these, and (2) Determining RNA folding transcriptome-wide in living organisms (plants, bacteria, and archaea) and evaluating implications of this folding on gene regulation and RNA processing. The former project has the advantage that one can precisely control experimental variable such as crowding, cosolutes, and ionic conditions on RNA folding, while the latter allows observations to be made made directly in a living organism.
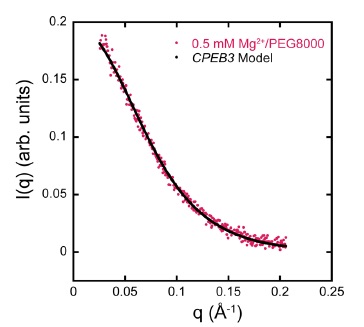 (1) RNA folding under in vivo-like conditions. One aspect of this project is to develop comprehensive molecular mechanisms for how functional RNAs fold in vivo and to relate these mechanisms to the evolutionary forces that help shape them. We have a longstanding interest in RNA folding. Presently, we are taking a comprehensive approach in which both the biophysical and evolutionary driving forces that give rise to RNA folding mechanism in vivo are identified. We are establishing biophysical principles for in vivo RNA folding by examining the folding mechanisms of several naturally occurring riboswitches and ribozymes in model cytoplasms. In addition we are elucidating evolutionary principles that guide RNA folding in vivo by testing the folding mechanisms of sequences that will emerge from several neutral drift selections. One recent advance has been determining that cellular-like crowding supports ribozyme reactivity by favoring a compact form of the ribozyme, but only under physiological ionic and cosolute conditions. SAXS studies are done in collaboration with Neela Yennawar [1]. Another study from the lab found that molecular crowders and cosolutes promote RNA folding cooperativity in physiological conditions [2].
(1) RNA folding under in vivo-like conditions. One aspect of this project is to develop comprehensive molecular mechanisms for how functional RNAs fold in vivo and to relate these mechanisms to the evolutionary forces that help shape them. We have a longstanding interest in RNA folding. Presently, we are taking a comprehensive approach in which both the biophysical and evolutionary driving forces that give rise to RNA folding mechanism in vivo are identified. We are establishing biophysical principles for in vivo RNA folding by examining the folding mechanisms of several naturally occurring riboswitches and ribozymes in model cytoplasms. In addition we are elucidating evolutionary principles that guide RNA folding in vivo by testing the folding mechanisms of sequences that will emerge from several neutral drift selections. One recent advance has been determining that cellular-like crowding supports ribozyme reactivity by favoring a compact form of the ribozyme, but only under physiological ionic and cosolute conditions. SAXS studies are done in collaboration with Neela Yennawar [1]. Another study from the lab found that molecular crowders and cosolutes promote RNA folding cooperativity in physiological conditions [2].
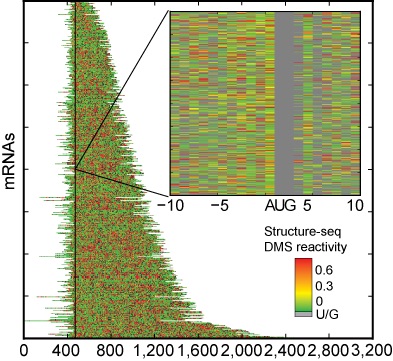 (2) RNA Folding Transcriptome-wide in Living Organisms. We recently developed a new approach to probe the structure of RNAs across an entire transcriptome in Arabidopsis [3] and a gene-specific way for low abundance RNAs [4]. In a related study, we demonstrated that RNA G-quadruplex downregulates translation of a DNA repair gene [5]. This work is funded by an NSF PGRP (plant genome research program) grant that is collaborative between our lab, Sally Assmann’s lab (Penn State Biology) on the plant biology, and David Mathews’ lab (U Rochester) on the computational implementation of structure prediction across a genome. Our goal in this work is to determine the folds of all the RNA in an entire transcriptome (tens of thousands of RNAs at the single nucleotide leve) in vivo and how they change during stress. We are identifying new paradigms for RNA folding in gene regulation. We are currently working with rice and Arabidopsis, but are moving into other organisms in the tree of life.
(2) RNA Folding Transcriptome-wide in Living Organisms. We recently developed a new approach to probe the structure of RNAs across an entire transcriptome in Arabidopsis [3] and a gene-specific way for low abundance RNAs [4]. In a related study, we demonstrated that RNA G-quadruplex downregulates translation of a DNA repair gene [5]. This work is funded by an NSF PGRP (plant genome research program) grant that is collaborative between our lab, Sally Assmann’s lab (Penn State Biology) on the plant biology, and David Mathews’ lab (U Rochester) on the computational implementation of structure prediction across a genome. Our goal in this work is to determine the folds of all the RNA in an entire transcriptome (tens of thousands of RNAs at the single nucleotide leve) in vivo and how they change during stress. We are identifying new paradigms for RNA folding in gene regulation. We are currently working with rice and Arabidopsis, but are moving into other organisms in the tree of life.Methods Applied
Thermodynamics: UV-detected thermal denaturations, ITC, DSC; Kinetics: Stopped-flow, T-jump; Structural: structure-mapping, SAXS; Genomics: Structure-seq, NGS, reporter-gene assays.
Group Members
Catherine Douds Elizabeth Jolley Jacob Sieg Megan Sylvia
Collaborators
Sally Assmann (Penn State University)
David Mathews (University of Rochester)
Neela Yennawar (Penn State University)
Recent Publications
- Yamagami, R., Kayedkhordeh, M., Mathews, D.H., Bevilacqua, P.C. Design of highly active double-pseudoknotted ribozymes: a combined computational and experimental study. Nucleic Acids Res. 47, 29-42 (2019) [PubMed] (open access).
- Mitchell III, D., Renda, A.J., Douds, C.A, Babitzke, P., Assmann, S.M., Bevilacqua, P.C. In vivo structural probing of uracil and guanine base pairing by 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) RNA 25, 147-157 (2019) [PubMed].
- Su, Z., Tang, Y., Ritchey, L.E., Tack, D.C., Zhu, M., Bevilacqua, P. C., Assmann, S. M. Genome-wide RNA structurome reprogramming reveals temperature-dependent regulation. Proc. Natl. Acad. Sci. 115, 12170-12175 (2018) [PubMed] (open access).
- Yamagami R, Bingaman JL, Frankel EA, Bevilacqua PC. Cellular conditions of weakly chelated magnesium ions strongly promote RNA stability and catalysis. Nature Communications 9, 2149 (2018) [PubMed] (open access).
- Leamy, K. A., Yennawar, N. H. & Bevilacqua, P. C. “Cooperative RNA folding under cellular conditions arises from both tertiary structure stabilization and secondary structure destabilization.” Biochemistry 56, 3422-3433 (2017). [PubMed] (link for 50 free e-prints)
Review Articles
- Mitchell, D., Assmann, S. M., Bevilacqua, P. C. Probing RNA structure in vivo. Curr. Opin. Struct. Biol. (in press).
- Bevilacqua, P.C. & Assmann, S.M. Technique development for probing RNA structure in vivo and genome-wide. Book Chapter in RNA World 5th Editors: J. Atkins, T.R. Cech, J. A. Steitz. (2018). [PubMed] (link to 50 free e-prints).
- Leamy, K. A., Assmann, S. M., Mathews, D. H., Bevilacqua, P. C.. “Bridging the gap between in vitro and in vivo RNA folding.” Q. Rev. Biophys. 49, e10 (2016). [PubMed]
- Bevilacqua, P. C., Ritchey, L. E., Su, Z., Assmann, S. M. “Genome-wide analysis of RNA secondary structure”. Annu. Rev. Genet. 50, 235-266 (2016). [PubMed]
- Kwok, C.K., Tang, Y., Assmann, S. M., Bevilacqua, P. C. “The RNA structurome: transcriptome-wide structure probing with next-gen sequencing” Trends Biochem. Sci. 40, 221-232 (2015). [PubMed]
